Sourcepredict example 1: Gut host species prediction¶
[1]:
import pandas as pd
import pandas_ml as pdml
In this example, we will use Sourcepredict and Sourcetracker2 applied to the example dataset provided in the Sourcepredict directory.
The example datasets contains the following samples: - Homo sapiens gut microbiome (1, 2, 3, 4, 5, 6) - Canis familiaris gut microbiome (1) - Soil microbiome (1, 2, 3)
Preparing the data¶
[2]:
cnt = pd.read_csv("../data/modern_gut_microbiomes_sources.csv", index_col=0)
labels = pd.read_csv("../data/modern_gut_microbiomes_labels.csv", index_col=0)
This is a TAXID count table containing the samples as columns headers, and the TAXID as row indices
[3]:
cnt.head()
[3]:
| SRR1175007 | SRR042182 | SRR061154 | SRR061499 | SRR063469 | SRR062324 | SRR1179037 | SRR061236 | SRR061456 | SRR642021 | ... | mgm4477903_3 | mgm4477807_3 | mgm4477874_3 | mgm4477904_3 | mgm4477804_3 | mgm4477873_3 | ERR1939166 | SRR3578625 | ERR1939165 | SRR3578645 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TAXID | |||||||||||||||||||||
| 0 | 3528337.0 | 11563613.0 | 10084261.0 | 20054993.0 | 8747525.0 | 12116517.0 | 4191329.0 | 13992760.0 | 14825759.0 | 11083673.0 | ... | 6169203.0 | 8820851.0 | 5713837.0 | 10238500.0 | 5055930.0 | 10380594.0 | 13391896.0 | 1553.0 | 14802198.0 | 736.0 |
| 6 | 0.0 | 78.0 | 0.0 | 127.0 | 0.0 | 79.0 | 0.0 | 0.0 | 0.0 | 172.0 | ... | 68.0 | 247.0 | 211.0 | 156.0 | 147.0 | 383.0 | 1353.0 | 0.0 | 1522.0 | 0.0 |
| 7 | 0.0 | 78.0 | 0.0 | 127.0 | 0.0 | 79.0 | 0.0 | 0.0 | 0.0 | 172.0 | ... | 68.0 | 247.0 | 211.0 | 156.0 | 147.0 | 383.0 | 1353.0 | 0.0 | 1522.0 | 0.0 |
| 9 | 0.0 | 129.0 | 0.0 | 153.0 | 0.0 | 151.0 | 0.0 | 165.0 | 96.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 77.0 | 0.0 | 65.0 | 0.0 |
| 10 | 0.0 | 160.0 | 0.0 | 193.0 | 0.0 | 99.0 | 0.0 | 55.0 | 249.0 | 238.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 263.0 | 0.0 | 466.0 | 0.0 |
5 rows × 432 columns
The labels file contains the mapping of samples names with their actual origin (sources)
[4]:
labels.head()
[4]:
| labels | |
|---|---|
| SRR1175007 | Homo_sapiens |
| SRR042182 | Homo_sapiens |
| SRR061154 | Homo_sapiens |
| SRR061499 | Homo_sapiens |
| SRR063469 | Homo_sapiens |
We will divide the source in training (95%) and testing (5%) dataset
[5]:
cnt_train = cnt.sample(frac=0.95, axis=1)
cnt_test = cnt.drop(cnt_train.columns, axis=1)
We also have to subset the labels file to only the training dataset
[6]:
train_labels = labels.loc[cnt_train.columns,:]
test_labels = labels.loc[cnt_test.columns,:]
Sourcepredict¶
Last but not least, we must export the files to csv to run sourcepredict
[7]:
cnt_train.to_csv("gut_species_sources.csv")
cnt_test.to_csv("gut_species_sinks.csv")
train_labels.to_csv("gut_species_labels.csv")
We’ll now launch sourcepredict with the GMPR normalization method, and the t-SNE embedding, on 6 cores.
[8]:
%%time
!sourcepredict -s gut_species_sources.csv \
-l gut_species_labels.csv \
-n GMPR \
-m TSNE \
-e example_embedding.csv \
-t 6 gut_species_sinks.csv
Step 1: Checking for unknown proportion
== Sample: SRR1175007 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR1175007
known:98.68%
unknown:1.32%
== Sample: SRR061236 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR061236
known:85.01%
unknown:14.99%
== Sample: SRR063471 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR063471
known:98.4%
unknown:1.6%
== Sample: SRR1930132 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR1930132
known:98.48%
unknown:1.52%
== Sample: SRR1930133 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.99
----------------------
- Sample: SRR1930133
known:98.49%
unknown:1.51%
== Sample: SRR7658586 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR7658586
known:79.65%
unknown:20.35%
== Sample: SRR7658645 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.99
----------------------
- Sample: SRR7658645
known:26.88%
unknown:73.12%
== Sample: SRR7658584 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.98
----------------------
- Sample: SRR7658584
known:85.78%
unknown:14.22%
== Sample: SRR7658607 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.99
----------------------
- Sample: SRR7658607
known:98.74%
unknown:1.26%
== Sample: SRR7658597 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.98
----------------------
- Sample: SRR7658597
known:99.1%
unknown:0.9%
== Sample: SRR5898944 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: SRR5898944
known:98.48%
unknown:1.52%
== Sample: ERR1914439 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1914439
known:98.48%
unknown:1.52%
== Sample: ERR1915140 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1915140
known:98.48%
unknown:1.52%
== Sample: ERR1914041 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1914041
known:98.48%
unknown:1.52%
== Sample: ERR1915022 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1915022
known:98.48%
unknown:1.52%
== Sample: ERR1915826 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1915826
known:98.48%
unknown:1.52%
== Sample: ERR1913400 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1913400
known:98.48%
unknown:1.52%
== Sample: ERR1915765 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1915765
known:98.48%
unknown:1.52%
== Sample: ERR1915225 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: ERR1915225
known:98.48%
unknown:1.52%
== Sample: mgm4477874_3 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 1.0
----------------------
- Sample: mgm4477874_3
known:72.37%
unknown:27.63%
== Sample: ERR1939166 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.97
----------------------
- Sample: ERR1939166
known:47.44%
unknown:52.56%
== Sample: ERR1939165 ==
Adding unknown
Normalizing (GMPR)
Computing Bray-Curtis distance
Performing MDS embedding in 2 dimensions
KNN machine learning
Training KNN classifier on 6 cores...
-> Testing Accuracy: 0.97
----------------------
- Sample: ERR1939165
known:55.27%
unknown:44.73%
Step 2: Checking for source proportion
Computing weighted_unifrac distance on species rank
TSNE embedding in 2 dimensions
KNN machine learning
Performing 5 fold cross validation on 6 cores...
Trained KNN classifier with 10 neighbors
-> Testing Accuracy: 0.99
----------------------
- Sample: SRR1175007
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR061236
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR063471
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR1930132
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR1930133
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR7658586
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR7658645
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR7658584
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR7658607
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR7658597
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: SRR5898944
Canis_familiaris:1.81%
Homo_sapiens:96.72%
Soil:1.47%
- Sample: ERR1914439
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1915140
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1914041
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1915022
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1915826
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1913400
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1915765
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: ERR1915225
Canis_familiaris:94.25%
Homo_sapiens:4.28%
Soil:1.47%
- Sample: mgm4477874_3
Canis_familiaris:2.51%
Homo_sapiens:5.95%
Soil:91.53%
- Sample: ERR1939166
Canis_familiaris:2.51%
Homo_sapiens:5.95%
Soil:91.53%
- Sample: ERR1939165
Canis_familiaris:2.51%
Homo_sapiens:5.95%
Soil:91.53%
Sourcepredict result written to gut_species_sinks.sourcepredict.csv
Embedding coordinates written to example_embedding.csv
CPU times: user 3.64 s, sys: 828 ms, total: 4.47 s
Wall time: 4min 2s
Two files were generated by Sourcepredict: - gut_species_sinks.sourcepredict.csv which contains the proportions of each source
[9]:
sourcepred = pd.read_csv("gut_species_sinks.sourcepredict.csv", index_col=0)
[10]:
sourcepred
[10]:
| SRR1175007 | SRR061236 | SRR063471 | SRR1930132 | SRR1930133 | SRR7658586 | SRR7658645 | SRR7658584 | SRR7658607 | SRR7658597 | ... | ERR1915140 | ERR1914041 | ERR1915022 | ERR1915826 | ERR1913400 | ERR1915765 | ERR1915225 | mgm4477874_3 | ERR1939166 | ERR1939165 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Canis_familiaris | 0.017814 | 0.015347 | 0.017765 | 0.017779 | 0.017781 | 0.014379 | 0.004853 | 0.015486 | 0.017825 | 0.017891 | ... | 0.928216 | 0.928216 | 0.928216 | 0.928216 | 0.928216 | 0.928216 | 0.928216 | 0.018200 | 0.011931 | 0.013898 |
| Homo_sapiens | 0.954443 | 0.822254 | 0.951788 | 0.952581 | 0.952644 | 0.770401 | 0.260037 | 0.829699 | 0.955022 | 0.958548 | ... | 0.042128 | 0.042128 | 0.042128 | 0.042128 | 0.042128 | 0.042128 | 0.042128 | 0.043078 | 0.028241 | 0.032896 |
| Soil | 0.014516 | 0.012506 | 0.014476 | 0.014488 | 0.014489 | 0.011717 | 0.003955 | 0.012619 | 0.014525 | 0.014579 | ... | 0.014504 | 0.014504 | 0.014504 | 0.014504 | 0.014504 | 0.014504 | 0.014504 | 0.662431 | 0.434275 | 0.505859 |
| unknown | 0.013226 | 0.149893 | 0.015971 | 0.015152 | 0.015086 | 0.203503 | 0.731155 | 0.142196 | 0.012627 | 0.008983 | ... | 0.015152 | 0.015152 | 0.015152 | 0.015152 | 0.015152 | 0.015152 | 0.015152 | 0.276292 | 0.525554 | 0.447347 |
4 rows × 22 columns
Let’s check which organism was predicted for each samples, and compare it with the true source
[11]:
comparison = sourcepred.idxmax().to_frame(name="prediction").merge(test_labels, left_index=True, right_index=True)
cm = pdml.ConfusionMatrix(y_true=comparison['labels'],y_pred=comparison['prediction'])
/Users/borry/miniconda3/lib/python3.6/site-packages/pandas/core/indexing.py:1494: FutureWarning:
Passing list-likes to .loc or [] with any missing label will raise
KeyError in the future, you can use .reindex() as an alternative.
See the documentation here:
https://pandas.pydata.org/pandas-docs/stable/indexing.html#deprecate-loc-reindex-listlike
return self._getitem_tuple(key)
Let’s look at the confusion matrix
[30]:
cm.to_dataframe()
[30]:
| Predicted | Canis_familiaris | Homo_sapiens | Soil | unknown |
|---|---|---|---|---|
| Actual | ||||
| Canis_familiaris | 8 | 0 | 0 | 0 |
| Homo_sapiens | 0 | 10 | 0 | 1 |
| Soil | 0 | 0 | 2 | 1 |
| unknown | 0 | 0 | 0 | 0 |
Finally, let’s compute the accuracy
[31]:
round(cm.stats()['overall']['Accuracy'],2)
[31]:
0.91
91% of the sink samples were correctly predicted !
- The second file generated by sourcepredict is
example_embedding.csvwhich contains the embedding coordinates of all samples (sources and sinks)
[14]:
embed = pd.read_csv("example_embedding.csv", index_col=0)
embed.head()
[14]:
| PC1 | PC2 | labels | name | |
|---|---|---|---|---|
| SRR1761672 | -12.706208 | -7.860738 | Homo_sapiens | SRR1761672 |
| SRR061456 | -4.520492 | 24.795073 | Homo_sapiens | SRR061456 |
| SRR1761718 | -20.427488 | -6.425568 | Homo_sapiens | SRR1761718 |
| SRR7658589 | -23.176891 | -2.985772 | Homo_sapiens | SRR7658589 |
| ERR1914932 | 28.669333 | 12.863045 | Canis_familiaris | ERR1914932 |
We can plot this embedding, using for example, plotnine, which implements the grammar of graphics in Python
[15]:
from plotnine import *
import warnings
warnings.filterwarnings('ignore')
[16]:
ggplot(data = embed, mapping = aes(x="PC1",y="PC2", color="labels")) + geom_point() + theme_classic()
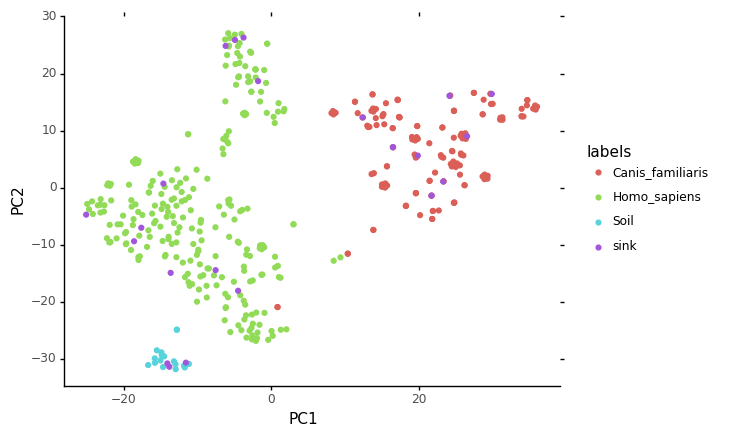
[16]:
<ggplot: (-9223372029842878797)>
We can see on this plot where the sink samples were embedded
Sourcetracker2¶
“SourceTracker is designed to predict the source of microbial communities in a set of input samples” and is generally consired as the gold standard method to do so. The version 2 is a rewrite of the original Sourcetracker in Python.
We’ll reuse the same training and test files, but we need to reformat them a bit for sourcetracker: - In sourcetracker, the source (training) and sink (file) TAXIDs count table is a single file - The metadata file is slightly different
[17]:
cnt.to_csv("gut_species_taxid.csv", sep="\t", index_label="TAXID")
[18]:
test_labels['SourceSink'] = ['sink']*test_labels.shape[0]
[19]:
train_labels['SourceSink'] = ['source']*train_labels.shape[0]
[20]:
metadata = train_labels.append(test_labels).rename(columns = {"labels":"Env"})[['SourceSink','Env']]
metadata.head()
[20]:
| SourceSink | Env | |
|---|---|---|
| SRR1761672 | source | Homo_sapiens |
| SRR061456 | source | Homo_sapiens |
| SRR1761718 | source | Homo_sapiens |
| SRR7658589 | source | Homo_sapiens |
| ERR1914932 | source | Canis_familiaris |
[21]:
metadata.to_csv("st_gut_species_metadata.csv", sep="\t", index_label='#SampleID')
Finally, we need to convert the TAXIDs count table to biom format
[22]:
!biom convert -i gut_species_taxid.csv -o gut_species_taxid.biom --table-type="Taxon table" --to-json
sourcetracker2 gibbs -i gut_species_taxid.biom -m st_gut_species_metadata.csv -o gut_species --jobs 6[32]:
st_pred = pd.read_csv("gut_species/mixing_proportions.txt", sep = "\t", index_col=0)
st_pred.head()
[32]:
| Canis_familiaris | Homo_sapiens | Soil | Unknown | |
|---|---|---|---|---|
| #SampleID | ||||
| SRR1175007 | 0.0170 | 0.9609 | 0.0063 | 0.0158 |
| SRR061236 | 0.0358 | 0.9365 | 0.0074 | 0.0203 |
| SRR063471 | 0.0121 | 0.9724 | 0.0032 | 0.0123 |
| SRR1930132 | 0.1466 | 0.3761 | 0.4477 | 0.0296 |
| SRR1930133 | 0.1182 | 0.5082 | 0.3507 | 0.0229 |
[34]:
st_comparison = st_pred.idxmax(axis=1).to_frame(name="prediction")
st_comparison.head()
[34]:
| prediction | |
|---|---|
| #SampleID | |
| SRR1175007 | Homo_sapiens |
| SRR061236 | Homo_sapiens |
| SRR063471 | Homo_sapiens |
| SRR1930132 | Soil |
| SRR1930133 | Homo_sapiens |
Let’s compare the SourceTracker prediction with the true source
[35]:
comparison2 = st_comparison.merge(test_labels, left_index=True, right_index=True)
comparison2
[35]:
| prediction | labels | SourceSink | |
|---|---|---|---|
| SRR1175007 | Homo_sapiens | Homo_sapiens | sink |
| SRR061236 | Homo_sapiens | Homo_sapiens | sink |
| SRR063471 | Homo_sapiens | Homo_sapiens | sink |
| SRR1930132 | Soil | Homo_sapiens | sink |
| SRR1930133 | Homo_sapiens | Homo_sapiens | sink |
| SRR7658586 | Homo_sapiens | Homo_sapiens | sink |
| SRR7658645 | Homo_sapiens | Homo_sapiens | sink |
| SRR7658584 | Soil | Homo_sapiens | sink |
| SRR7658607 | Homo_sapiens | Homo_sapiens | sink |
| SRR7658597 | Homo_sapiens | Homo_sapiens | sink |
| SRR5898944 | Homo_sapiens | Homo_sapiens | sink |
| ERR1914439 | Canis_familiaris | Canis_familiaris | sink |
| ERR1915140 | Soil | Canis_familiaris | sink |
| ERR1914041 | Canis_familiaris | Canis_familiaris | sink |
| ERR1915022 | Canis_familiaris | Canis_familiaris | sink |
| ERR1915826 | Canis_familiaris | Canis_familiaris | sink |
| ERR1913400 | Canis_familiaris | Canis_familiaris | sink |
| ERR1915765 | Canis_familiaris | Canis_familiaris | sink |
| ERR1915225 | Canis_familiaris | Canis_familiaris | sink |
| mgm4477874_3 | Soil | Soil | sink |
| ERR1939166 | Soil | Soil | sink |
| ERR1939165 | Soil | Soil | sink |
Computing the accuracy
[36]:
cm2 = pdml.ConfusionMatrix(y_true=comparison2["labels"], y_pred=comparison2["prediction"])
cm2.to_dataframe()
[36]:
| Predicted | Canis_familiaris | Homo_sapiens | Soil |
|---|---|---|---|
| Actual | |||
| Canis_familiaris | 7 | 0 | 1 |
| Homo_sapiens | 0 | 9 | 2 |
| Soil | 0 | 0 | 3 |
[38]:
acc2 = round(cm2.stats()['overall']['Accuracy'],2)
[38]:
0.86
Here, Sourcetracker only managed to predict 86% of the sink samples origin correctly
Conclusion¶
On this dataset, we’ve seen that Sourcepredict performs similary or even better than Sourcetracker on predicting accurately the source species